This tutorial describes the general approach of phylogenetic tree construction using RAxML, focusing on a example using the thioredoxin gene family in Arabidopsis thaliana. The analysis will incorporate the use of domain identification, domain extraction, multiple sequence alignment, phylogenetic tree construction, bootstrap analysis, and biological interpretation.
Examples files used in this analysis can be found on Github in the examples folder of QKdomain.
Identification of thioredoxins in Arabidopsis thaliana
The thioredoxins of Arabidopsis thaliana were identified by identifying all proteins with the Pfam domain PF00085. Next, InterProScan was used to evaluate these sequences. The resulting output in tab-separated format (tsv) is used by the QKdomain (v1.0) suite of scripts to identify the diversity of domains. Initially, QKdomain_preprocess.py is used to assess the domain content present in the protein data set.
python QKdomain_preprocess.py Athaliana_167_TAIR10.protein.TRX.fa.tsv At_TRX_preprocess.txt
The output file At_TRX_preprocess.txt is used to manually curate identifiers to generate redundant abbreviations for the next component of the analysis. Below is an example of a set of abbreviations generated from At_TRX_preprocess.txt.
Coil CC
PF00085 TRX
PF00462 GLRX
PF00515 TPR
PF00789 UBX
...
Next, we identify the domain structure of proteins in the data set.
python QKdomain_process.py Athaliana_167_TAIR10.protein.TRX.fa Athaliana_167_TAIR10.protein.TRX.fa.tsv At_TRX_abbreviations.txt At_TRX_process.txt
The output will be structured with the first column the protein identifier and the second column the protein domain structure.
AT1G35620.1 TRX-TRX
AT3G56420.1 TRX
AT5G66410.1 TRX
AT1G14570.1 UBA-TRX-UIM-UBX-UBI
AT3G53220.1 TRX
AT3G15360.1 TRX
AT2G32920.1 TRX-TRX-TRX
AT1G08570.1 CC-TRX
...
To export all thioredoxin (TRX) domains in the set of proteins, use the following command:
python QKdomain_process.py -d TRX Athaliana_167_TAIR10.protein.TRX.fa Athaliana_167_TAIR10.protein.TRX.fa.tsv At_TRX_abbreviations.txt At_TRX_process.txt At.TRX.domain.fa
The file At.TRX.domain.fa will contain individual non-overlapping domains delineated by InterProScan. Overlapping domains would be integrated into a single domain and would require manual curation in a multiple sequence alignment to correct. The protein identifier is modified to include the start and end positions selected and the domain structure of the original protein.
Multiple sequence alignment of thioredoxin domains from Arabidopsis thaliana
A multiple sequence alignment is critical for the development of an accurate phylogenetic tree. Several forms of software are available, and each has different strengths according the the size and complexity of the data set under investigation. A summary table of commonly used software is listed below (Table 1). A considerable number of additional options are available and this list should just be considering a starting point. In addition, parallel versions exist for several software, such as ClustalOmega and MUSCLE.
Table 1. Multiple sequence alignment software
| Software | Scalability | Complexity | Website |
|---|---|---|---|
| ClustalW | Small to Medium | Diverse sequences (more conservative) | http://www.clustal.org/clustal2/ |
| ClustalOmega | Large | Diverse sequences (more relaxed) | http://www.clustal.org/omega/ |
| MAFFT | Medium to Large | http://mafft.cbrc.jp/alignment/software/ | |
| MUSCLE | Medium | Diverse sequences (more conservative) | http://www.drive5.com/muscle/ |
| PRANK | Medium | Low complexity (highly related sequences) | http://wasabiapp.org/software/prank/ |
The thioredoxins of Arabidopsis thaliana are fairly diverse, therefore MUSCLE was selected for multiple sequence alignment. The first step is to identify a conserved region in the thioredoxin domain and removal of sequences unrelated to the thioredoxin domain and/or without conservation in this gene family.
muscle -in At.TRX.domain.fa -clwstrict -out At.TRX.domain.MUSCLE1.aln
A clear cutoff can be identified between positions 382 to 581 aa in the alignment. Next, we realign the extracted sequence, which can improve the assembly once divergent sequences are removed.
muscle -in At.TRX.domain.MUSCLE1_382_581.fa -clwstrict -out At.TRX.domain.MUSCLE2.aln
While the first alignment and selection will remove the majority of outlier sequence, it is important to check and remove any additional sequence. In this case, an extraction was made between the positions 34 and 217 aa in the multiple sequence alignment.
muscle -in At.TRX.domain.MUSCLE2_34_217.fa -clwstrict -out At.TRX.domain.MUSCLE3.aln
The last step involves an iterative process of removing sequences that are highly divergent from the other sequences in the alignment. This is performed by (1) performing the multiple sequence alignment, (2) construction of a neighbor joining tree, and (3) identification of genes that are highly divergent based on the tree structure. The details of the removed sequences are detailed below (Table 2).
muscle -in At.TRX.domain.MUSCLE2_34_217_curated.fa -clwstrict -out At.TRX.domain.MUSCLEf.aln
Table 2. Manual curation of sequences in multiple sequence alignment based on aberrant phylogenetic inference.
| Iteration | Gene |
|---|---|
| 1 | AT1G20225.1_8_210 |
| 1 | AT1G35620.1_180_327 |
| 2 | AT4G04950.1_153_256 |
| 3 | AT1G60420.1_188_299 |
| 3 | AT3G54960.2_208_518 |
| 4 | AT4G14250.1_500_624 |
| 4 | AT4G14250.2_105_191 |
An example of how the tree is incorrect is seen below.

After removal of divergent sequence, the tree takes its correct structure.
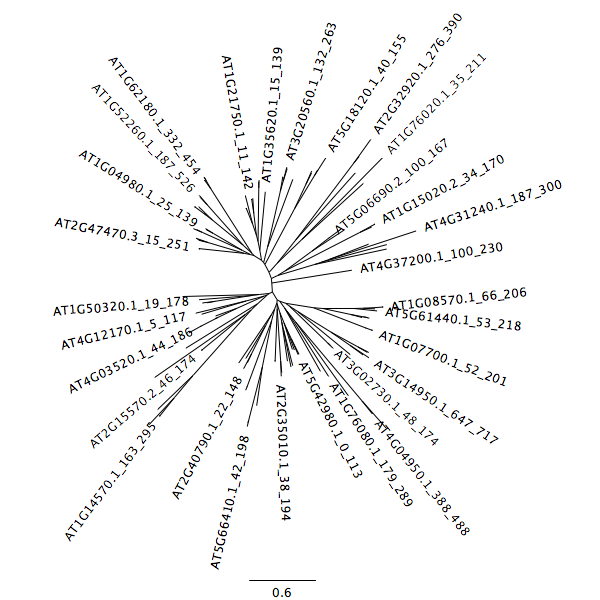
Phylogenetic tree construction of thioredoxin domains from Arabidopsis thaliana
Several approaches and software are available for phylogenetic tree construction including but not limited to parsimony, neighbor-joining, and maximum likelihood. Here, we demonstrate how to generate a maximum likelihood phylogenetic tree using RAxML. First, we generate the phylogenetic tree. In the first instance, we will use the automatic selection of amino acid selection model (PROTGAMMAAUTO) and use four processors -T 4.
raxml -s At.TRX.domain.MUSCLEf.phy -n At.TRX.domain.PROTGAMMAAUTO -m PROTGAMMAAUTO -p 84381764921 -T 4
The output will initially describe the set of sequences that are identical such as:
IMPORTANT WARNING: Sequences AT1G14570.1_163_295 and AT1G14570.3_142_274 are exactly identical
IMPORTANT WARNING: Sequences AT2G15570.2_46_174 and AT2G15570.1_46_173 are exactly identical
IMPORTANT WARNING: Sequences AT1G07700.3_65_214 and AT1G07700.1_52_201 are exactly identical
It will then perform automatic protein model assignment until it identifies an appropriate model. In this case, WAG. The final output will be in the file RAxML_result.At.TRX.domain.PROTGAMMAAUTO. The run time was 443 seconds using four processors on a relatively new MacBook Pro. This is important, as the length of the alignment and number of sequences will have a tremendous impact on the time required for phylogenetic tree construction. We routinely use large multicore servers to handle large data sets.
Next, we need to generate bootstraps to infer relationships of genes within the tree. This will take a considerable amount of time, but we can test for convergence as we perform blocks of bootstrap analyses.
raxml -s At.TRX.domain.MUSCLEf.phy -n At.TRX.domain.PROTGAMMAAUTO_bootstrap_r1 -N 100 -m PROTGAMMAWAG -p 427482396541 -T 4
raxml -s At.TRX.domain.MUSCLEf.phy -n At.TRX.domain.PROTGAMMAAUTO_bootstrap_r2 -N 100 -m PROTGAMMAWAG -p 9567154133 -T 4
cat RAxML_result.At.TRX.domain.PROTGAMMAAUTO_bootstrap_r* > allBootstraps
raxml -z allBootstraps -m PROTGAMMAWAG -I autoMRE -n TEST -p 3824142315 -T 4
On my machine, it tooks 5.03 hours to run the first 100 bootstraps. For testing this tutorial on your own computer, I would suggest preparing a small data set in the first instance. After 10 hours of running, the convergence test was met after 200 bootstraps. To incorporate bootstraps into the maxmimum likelihood tree, we used the following command:
raxml -f b -z allBootstraps -t RAxML_result.At.TRX.domain.PROTGAMMAAUTO -m PROTGAMMAWAG -n At.TRX.domain.PROTGAMMAWAGandBOOTSTRAP.txt
The file At.TRX.domain.PROTGAMMAWAGandBOOTSTRAP.txt can be directly used with EMBL iTOL for visualization.
Files used in all analyses
All files used in this analysis can be accessed on my webpage source account on Github.