Research
Our group focuses on understanding immunity in the grasses

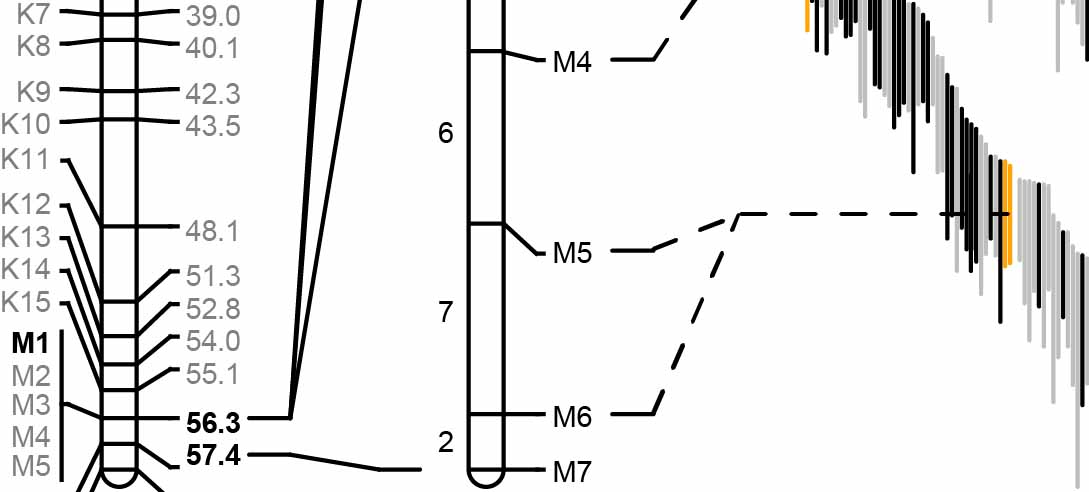
Genetic architecture of nonhost resistance
Identifying nonhost resistance genes that maintain immunity to cereal rusts (Puccinia spp.)
Tradeoffs between biotrophs and necrotrophs
The molecular tradeoffs of R genes between resistance to biotrophs and susceptibility to necrotrophs

Genomics of immune receptors in the grasses
Defining the set of immune receptors in the grasses and understanding the evolution of immunity